Creating Custom Albumentations Transforms 🔗
While Albumentations provides a wide array of built-in transformations, you might need to create your own custom logic. This guide walks through the process step-by-step, referencing the typical flow of a custom transform.
Motivation 🔗
Why create a custom transform?
- Implement novel augmentation techniques.
- Create domain-specific transforms.
- Wrap functions from external image processing libraries.
- Encapsulate complex or conditional logic.
Quick Reference: Transform Creation Essentials 🔗
Learning Path 🔗
- 🟢 Essential: Steps 1-2 (Image-only transforms)
- 🟡 Intermediate: Step 3 (Multiple images)
- 🟠 Advanced: Step 4 (Multi-target), 🟠 Validation, 🟠 Custom Data
Key Concepts 🔗
# Base Classes (choose one)
A.ImageOnlyTransform # Modifies only image pixels
A.DualTransform # Modifies image + mask/bboxes/keypoints
A.Transform3D # For 3D/volumetric data
# Essential Methods (implement these)
def __init__(self, p=0.5): # Setup & parameters
def get_params_dependent_on_data(self, params, data): # Sample random values
def apply(self, img, **params): # Transform logic
Critical Rules 🔗
- ⚠️ Probability Default:
p=0.5by default - usep=1.0for always-applied transforms - 🎲 Reproducibility: Use
self.py_randomorself.random_generator(never globalrandom) - 📊 Parameter Sampling: Do ALL random sampling in
get_params_dependent_on_data
Transform Types Quick Guide 🔗
| Use Case | Base Class | Key Methods |
|---|---|---|
| Change pixels only | ImageOnlyTransform | apply |
| Change image + mask/bboxes/keypoints | DualTransform | apply, apply_to_mask, apply_to_bboxes, apply_to_keypoints |
| Handle 3D volumes | Transform3D | apply_to_volume |
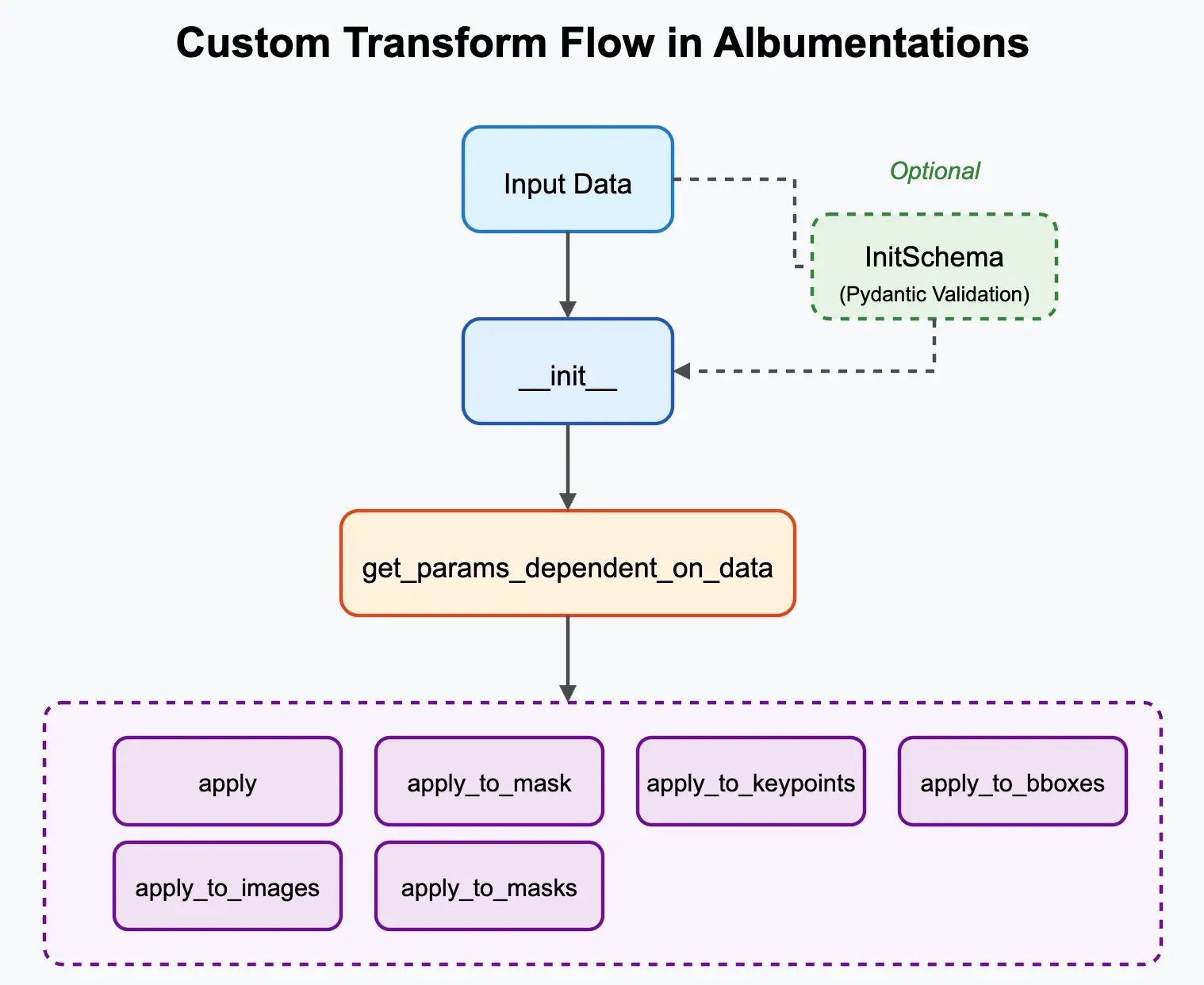
Core Concept: Inheriting from Base Classes 🔗
To integrate with Albumentations, custom transforms inherit from base classes like A.ImageOnlyTransform, A.DualTransform, or A.Transform3D. These base classes provide the structure for handling different data targets (image, mask, bboxes, etc.) and probabilities.
⚠️ Important: Default Probability 🔗
The BasicTransform class, which all transform classes inherit from, has a default value of p=0.5. This means if you don't explicitly specify the probability parameter in your custom transform's constructor, it will only be applied 50% of the time.
If you want your transform to be always applied, make sure to pass p=1.0 to the parent class constructor:
class MyTransform(A.ImageOnlyTransform):
def __init__(self):
# This transform will only be applied 50% of the time
super().__init__() # p defaults to 0.5
class MyAlwaysAppliedTransform(A.ImageOnlyTransform):
def __init__(self):
# This transform will always be applied
super().__init__(p=1.0)
🟢 Step 1: Minimal Image-Only Transform (Essential) 🔗
Let's start with the simplest case: a transform that only modifies the image pixels and involves some randomness. We'll inherit from A.ImageOnlyTransform.
Goal: Create a transform that multiplies the image pixel values by a random factor between 0.5 and 1.5.
import albumentations as A
from albumentations.core.transforms_interface import ImageOnlyTransform
import numpy as np
import random
from typing import Any
class RandomMultiplier(ImageOnlyTransform):
"""Multiplies the pixel values of an image by a random factor."""
def __init__(self, factor_range=(0.5, 1.5), p=0.5):
# 1. Initialize the base class with probability 'p'
super().__init__(p=p)
# Store initialization parameters
self.factor_range = factor_range
# Use get_params_dependent_on_data for ALL parameter sampling
# Correct signature includes `params` from get_params (though unused here)
def get_params_dependent_on_data(self, params, data):
# 2. Sample random parameter(s) once per call.
# Even if not strictly dependent, we use this method.
# The 'data' dict (e.g., data['image']) is available here if needed.
factor = random.uniform(self.factor_range[0], self.factor_range[1])
return {"factor": factor}
def apply(self, img, factor, **params):
# 3. Apply the transformation logic to the image
# 'factor' comes from the dictionary returned by get_params_dependent_on_data()
# We use np.clip to keep values in the valid range [0, 255]
return np.clip(img * factor, 0, 255).astype(img.dtype)
# --- Usage ---
# Instantiate the custom transform
random_mult = RandomMultiplier(factor_range=(0.8, 1.2), p=1.0)
# Use it in a pipeline
pipeline = A.Compose([
A.Resize(256, 256), # Example built-in transform
random_mult, # Your custom transform
])
Key Points:
__init__(self, ...): The constructor receives configuration parameters (factor_range). It callssuper().__init__(p=p)to handle the standard probability logic.get_params_dependent_on_data(self, params, data): This method is called once per transform invocation (if probabilitypis met). Its primary role is to sample any random values needed for the augmentation and perform any calculations (potentially based on the inputdatadictionary) that need to be shared across differentapply_to_...methods (likeapply,apply_to_mask, etc.). It receives theparamsdict fromget_params(which is empty in this workflow) and thedatadict. Returning values in a dictionary ensures they are computed only once and then passed consistently to the relevant apply methods via the**paramsargument.apply(self, img, factor, **params): Performs the transformation using the image (img) and the sampledfactor(received via**params). Returns the modified image.
This minimal example demonstrates the basic structure. get_params_dependent_on_data() is the method used for sampling augmentation parameters and centralizing calculations.
🟢 Step 2: Data-Dependent Image-Only Transform (Essential) 🔗
Now, let's create a transform where the parameters calculated depend on the input image itself. We'll still use ImageOnlyTransform as we only modify the image pixels.
Goal: Create a transform that normalizes the image using its per-channel mean and standard deviation.
import albumentations as A
from albumentations.core.transforms_interface import ImageOnlyTransform
import numpy as np
class PerImageNormalize(ImageOnlyTransform):
"""Normalizes an image using its own mean and standard deviation."""
def __init__(self, p=1.0):
# Only probability needed for initialization
super().__init__(p=p)
# Correct signature includes `params` from get_params (though unused here)
def get_params_dependent_on_data(self, params, data):
# Calculate mean and std from the input image
img = data['image'] # Access the image from the input data dict
mean = np.mean(img, axis=(0, 1))
std = np.std(img, axis=(0, 1))
# Add a small epsilon to std to avoid division by zero
std = np.where(std < 1e-6, 1e-6, std)
return {"mean": mean, "std": std}
def apply(self, img, mean, std, **params):
# Apply normalization using the calculated mean and std
# Ensure calculations are done in float to avoid data type issues
img = img.astype(np.float32)
normalized_img = (img - mean) / std
return normalized_img
# Note: Output dtype will be float32 after normalization
Explanation:
__init__: Only requires the probabilityp.get_params_dependent_on_data(self, params, data):- It receives the
datadictionary. We access the input image usingdata['image']. - We calculate the mean and standard deviation across the height and width dimensions (axes 0 and 1) for each channel.
- A small epsilon is added to the standard deviation to prevent division by zero in case of a completely flat channel.
- The calculated
meanandstdare returned in the parameters dictionary.
- It receives the
apply(self, img, mean, std, **params):- Receives the image and the calculated
meanandstdviaparams. - Performs the standard normalization formula:
(pixel - mean) / std. - The image is cast to
float32before the operation to ensure correct calculation and handle potential negative values after mean subtraction. The output will befloat32.
- Receives the image and the calculated
This example shows how get_params_dependent_on_data allows you to compute augmentation parameters based on the specific input image being processed.
🟡 Step 3: Handling Multiple Images Efficiently (Intermediate) 🔗
While ImageOnlyTransform focuses on modifying only image pixels, it can still receive multiple images via the images keyword argument (e.g., for video frames or batches needing consistent parameters). The default behavior might loop through each image slice and apply the single-image apply method using the same parameters for all slices.
For our PerImageNormalize example, this default behavior would be inefficient and incorrect (using the same mean/std for all images). We can override the apply_to_images method within our ImageOnlyTransform subclass to provide a correct and efficient vectorized implementation.
Goal: Efficiently normalize each image in a sequence/batch using its own mean and standard deviation, while still inheriting from ImageOnlyTransform.
import albumentations as A
from albumentations.core.transforms_interface import ImageOnlyTransform # Keep ImageOnlyTransform
import numpy as np
from typing import Any # For type hinting
class PerImageNormalizeEfficient(ImageOnlyTransform): # Renamed for clarity
"""
Normalizes an image or a sequence/batch of images using per-image
mean and standard deviation. Overrides apply_to_images for efficiency.
"""
def __init__(self, p=1.0):
super().__init__(p=p)
# get_params_dependent_on_data needs to handle both single image and multiple images
def get_params_dependent_on_data(self, params, data):
# Rename stats to params for clarity
params_out = {}
if 'image' in data:
img = data['image']
params_out['mean'] = np.mean(img, axis=(0, 1))
params_out['std'] = np.std(img, axis=(0, 1))
elif 'images' in data:
imgs = data['images'] # Shape (N, H, W, C) or (N, H, W)
# Calculate per-image mean/std. Axis is (1, 2) for H, W
params_out['mean'] = np.mean(imgs, axis=(1, 2))
params_out['std'] = np.std(imgs, axis=(1, 2))
# Ensure means/stds have shape (N, C) or (N,) for broadcasting
if imgs.ndim == 4 and params_out['mean'].ndim == 1: # Add channel dim if needed
params_out['mean'] = params_out['mean'][:, np.newaxis]
params_out['std'] = params_out['std'][:, np.newaxis]
else:
return {}
# Add epsilon for stability
params_out['std'] = np.where(params_out['std'] < 1e-6, 1e-6, params_out['std'])
return params_out # Return the parameters dictionary
# apply for single image (same as Step 2)
def apply(self, img, mean, std, **params):
# Ignore mode if necessary, or use for assertion/logging
img = img.astype(np.float32)
normalized_img = (img - mean) / std
return normalized_img
# Override apply_to_images for sequence/batch (vectorized)
def apply_to_images(self, images, mean, std, **params):
# Ignore mode if necessary, or use for assertion/logging
# Ensure calculations are done in float
images = images.astype(np.float32)
# mean and std have shape (N, C) or (N,) allowing broadcasting
# images shape is (N, H, W, C) or (N, H, W)
# Reshape mean/std for broadcasting: (N, 1, 1, C) or (N, 1, 1)
if images.ndim == 4: # N, H, W, C
mean = mean[:, np.newaxis, np.newaxis, :]
std = std[:, np.newaxis, np.newaxis, :]
elif images.ndim == 3: # N, H, W
mean = mean[:, np.newaxis, np.newaxis]
std = std[:, np.newaxis, np.newaxis]
normalized_images = (images - mean) / std
return normalized_images
# targets property is implicitly {"image": apply} from ImageOnlyTransform,
# but the pipeline logic will correctly call apply_to_images when the 'images'
# key is present in the input data.
Key Points:
- Inheritance: We still inherit from
A.ImageOnlyTransform. get_params_dependent_on_data: This is updated to detect if'image'or'images'is present in the inputdataand calculate the mean/std accordingly (either a single value/vector or an array of values/vectors). It returns the calculated parameters in a dictionary.apply: Handles the single-image case as before. It receives the singlemeanandstd.apply_to_images: This method is overridden. It receives theimagesstack/sequence and the arrays ofmeanandstdcalculated inget_params_dependent_on_data. It uses NumPy broadcasting to efficiently normalize each image with its corresponding statistics.
This PerImageNormalizeEfficient class now correctly handles both single images (via apply) and sequences/batches (via the overridden apply_to_images) while still being an ImageOnlyTransform, providing efficiency and correctness for the per-image normalization task.
🟠 Step 4: Geometric Transform Affecting Multiple Targets (Advanced) 🔗
This step shows how to implement a custom geometric transform inheriting from A.DualTransform and manually applying the transformation logic. Remember that apply_to_bboxes and apply_to_keypoints receive data in a standardized internal format.
Goal: Create a transform that randomly shifts the image and all associated targets, implementing the target logic manually.
import albumentations as A
from albumentations.core.transforms_interface import DualTransform
import numpy as np
import cv2
import random
from typing import Any, Sequence, Dict # Use Dict for type hint clarity
class RandomShiftMultiTargetManual(DualTransform):
"""
Randomly shifts the image and associated targets (mask, bboxes, keypoints)
with manual implementation of target transformations, respecting internal formats.
Processes arrays of bboxes and keypoints.
"""
def __init__(self, x_limit=0.1, y_limit=0.1,
interpolation=cv2.INTER_LINEAR,
mask_interpolation=cv2.INTER_NEAREST,
border_mode=cv2.BORDER_CONSTANT,
fill=0,
fill_mask=0,
p=0.5):
super().__init__(p=p)
self.x_limit = x_limit
self.y_limit = y_limit
self.interpolation = interpolation
self.mask_interpolation = mask_interpolation
self.border_mode = border_mode
self.fill = fill
self.fill_mask = fill_mask
def get_params_dependent_on_data(self, params, data):
height, width = data['image'].shape[:2]
# CORRECT: Use self.py_random for reproducibility
dx = self.py_random.uniform(-self.x_limit, self.x_limit)
dy = self.py_random.uniform(-self.y_limit, self.y_limit)
# INCORRECT (would ignore Albumentations seed):
# dx = random.uniform(-self.x_limit, self.x_limit)
# dy = random.uniform(-self.y_limit, self.y_limit)
# # Example using numpy generator if needed for array sampling:
# noise_level = self.random_generator.uniform(0.01, 0.05)
x_shift = int(width * dx)
y_shift = int(height * dy)
matrix = np.float32([[1, 0, x_shift], [0, 1, y_shift]])
return {
"matrix": matrix,
"x_shift": x_shift,
"y_shift": y_shift,
"height": height,
"width": width
}
def apply(self, img: np.ndarray, matrix: np.ndarray, height: int, width: int, **params):
return cv2.warpAffine(
img, matrix, (width, height),
flags=self.interpolation, borderMode=self.border_mode, borderValue=self.fill
)
def apply_to_mask(self, mask: np.ndarray, matrix: np.ndarray, height: int, width: int, **params):
return cv2.warpAffine(
mask, matrix, (width, height),
flags=self.mask_interpolation, borderMode=self.border_mode, borderValue=self.fill_mask
)
# Correct method name and signature for multiple bounding boxes
def apply_to_bboxes(self, bboxes: np.ndarray, x_shift: int, y_shift: int, height: int, width: int, **params):
"""
Applies shift to an array of bounding boxes.
Assumes bboxes array is in internal normalized format [:, [x_min, y_min, x_max, y_max, ...]].
Returns bboxes array in the same format.
"""
# Calculate normalized shifts
norm_dx = x_shift / width
norm_dy = y_shift / height
# Apply shifts vectorized
bboxes_shifted = bboxes.copy()
bboxes_shifted[:, [0, 2]] = bboxes[:, [0, 2]] + norm_dx # Shift x_min, x_max
bboxes_shifted[:, [1, 3]] = bboxes[:, [1, 3]] + norm_dy # Shift y_min, y_max
return bboxes_shifted
# Correct method name and signature for multiple keypoints
def apply_to_keypoints(self, keypoints: np.ndarray, x_shift: int, y_shift: int, **params):
"""
Applies shift to an array of keypoints.
Assumes keypoints array is in internal format [:, [x, y, angle, scale, ...]] where x, y are absolute pixels.
Returns keypoints array in the same format.
"""
keypoints_shifted = keypoints.copy()
keypoints_shifted[:, 0] = keypoints[:, 0] + x_shift # Shift x
keypoints_shifted[:, 1] = keypoints[:, 1] + y_shift # Shift y
# Angle and scale remain unchanged by translation
return keypoints_shifted
Using Custom Transforms in Pipelines 🔗
Once defined, your custom transform class is used just like any built-in Albumentations transform. Simply instantiate it and add it to your A.Compose list.
🔴 Advanced Topics 🔗
Reproducibility and Random Number Generation 🔗
To ensure your custom transforms produce reproducible results when a seed is set for the overall pipeline using A.Compose(..., seed=...), you must use the random number generators provided by the base transform classes: self.py_random and self.random_generator.
Using Python's global random module or NumPy's global np.random directly within your transform methods will bypass the seeding mechanism managed by Albumentations, leading to non-reproducible results even if a seed is provided to Compose. Fixing global seeds outside the pipeline also does not guarantee reproducibility within the pipeline's transformations.
The only way to ensure seeded reproducibility for random operations within your custom transform is to use the generators provided on self:
-
self.py_random:- An instance of Python's
random.Random, seeded based on theComposeseed. - Use for standard Python random functions (e.g.,
self.py_random.uniform(a, b)).
- An instance of Python's
-
self.random_generator:- An instance of NumPy's
numpy.random.Generator, seeded based on theComposeseed. - Use for NumPy-based random sampling (e.g.,
self.random_generator.uniform(a, b, size)).
- An instance of NumPy's
Reference RandomShiftMultiTargetManual.get_params_dependent_on_data: See the implementation in Step 4 for an example of correctly using self.py_random.uniform.
Always use self.py_random or self.random_generator within your get_params_dependent_on_data method for any random sampling.
🟠 Input Parameter Validation with InitSchema 🔗
When creating custom transforms, especially ones with multiple configuration parameters, it's crucial to validate the inputs provided during initialization (__init__). Without validation, incorrect parameter values (e.g., wrong type, out-of-range numbers) might only cause errors much later during the training loop, potentially deep into an epoch, making debugging difficult. This violates the "fail fast" principle.
Manually writing comprehensive validation logic (e.g., using if/else checks and raise ValueError) within the __init__ method for every parameter can be tedious, verbose, and error-prone. Developers might skip validating edge cases, leading to unexpected behavior later.
To address this, Albumentations integrates with the Pydantic library, providing a declarative and robust way to define input schemas and validation rules for your transform's __init__ parameters. This ensures parameters are checked thoroughly upon instantiation.
- Install Pydantic: If you don't have it:
pip install pydantic - Define
InitSchema: Inside your transform class, define an inner class namedInitSchemathat inherits frompydantic.BaseModel. - Declare Fields: Define the fields in
InitSchemacorresponding to your__init__arguments, using Pydantic's type hints andFieldfor basic constraints (likege,le). - (Optional) Add Custom Validators: Use Pydantic decorators like
@field_validator(for single fields) and@model_validator(for checks involving multiple fields) to implement more complex validation logic.
Albumentations will automatically find and use this InitSchema to validate the arguments when your transform is instantiated.
Example: Mutually Exclusive Parameters
Let's create a hypothetical CustomBlur transform that can apply either a Gaussian blur (requiring sigma) or a Box blur (requiring kernel_size), but not both.
import albumentations as A
from albumentations.core.transforms_interface import ImageOnlyTransform
import numpy as np
import cv2
from typing import Any, Sequence, Dict, Optional # Optional needed
# Requires: pip install pydantic
from pydantic import BaseModel, Field, model_validator, ValidationError
class CustomBlur(ImageOnlyTransform):
# Define Pydantic schema with custom model validator
class InitSchema(BaseModel):
sigma: float | None = Field(ge=0, description="Sigma for Gaussian blur.")
kernel_size: int | None = Field(ge=3, description="Kernel size for Box blur (must be odd).")
# Model validator to check mutual exclusivity and kernel_size oddness
@model_validator(mode='after')
def check_blur_params(self):
sigma_set = self.sigma is not None
kernel_set = self.kernel_size is not None
if sigma_set and kernel_set:
raise ValueError("Specify either 'sigma' or 'kernel_size', not both.")
if not sigma_set and not kernel_set:
raise ValueError("Must specify either 'sigma' (for Gaussian) or 'kernel_size' (for Box).")
if kernel_set and self.kernel_size % 2 == 0:
raise ValueError(f"kernel_size must be odd, got {self.kernel_size}")
return self
def __init__(self, sigma: float | None = None, kernel_size: int | None = None, p=0.5):
super().__init__(p=p)
# Store validated parameters (validation happens before __init__ fully runs)
self.sigma = sigma
self.kernel_size = kernel_size
def apply(self, img: np.ndarray, **params):
if self.sigma is not None:
# Ensure sigma is > 0 for cv2.GaussianBlur
sigma_safe = max(self.sigma, 1e-6)
return cv2.GaussianBlur(img, ksize=(0, 0), sigmaX=sigma_safe)
elif self.kernel_size is not None:
return cv2.blur(img, ksize=(self.kernel_size, self.kernel_size))
else:
# Should not be reached due to validation, but good practice
return img
# Since params aren't changing randomly per call, get_params_... is not needed
# --- Usage with Validation ---
try:
# Valid: Gaussian
transform_gauss = CustomBlur(sigma=1.5, p=1.0)
print("Gaussian blur instantiation OK.")
# Valid: Box blur
transform_box = CustomBlur(kernel_size=5, p=1.0)
print("Box blur instantiation OK.")
except ValidationError as e:
print(f"Unexpected validation error: {e}")
try:
# Fails: Both specified
transform_both = CustomBlur(sigma=1.5, kernel_size=5)
except ValidationError as e:
print(f"Instantiation failed (Both Params):\n{e}\n")
try:
# Fails: Neither specified
transform_neither = CustomBlur()
except ValidationError as e:
print(f"Instantiation failed (Neither Param):\n{e}\n")
try:
# Fails: Even kernel_size
transform_even_kernel = CustomBlur(kernel_size=4)
except ValidationError as e:
print(f"Instantiation failed (Even Kernel):\n{e}\n")
This example demonstrates how @model_validator enforces complex rules involving multiple __init__ parameters, ensuring the transform is configured correctly.
🟠 Passing Arbitrary Data via targets_as_params 🔗
Standard Albumentations targets like image, mask, bboxes, keypoints are automatically handled by the base transform classes (ImageOnlyTransform, DualTransform, etc.). However, sometimes you need to pass arbitrary data through the pipeline to your custom transform – data that isn't one of the standard types but is needed to determine the augmentation parameters.
Examples include:
- Overlay images/masks to be blended onto the main image.
- Metadata associated with the image (e.g., timestamps, sensor readings).
- Paths to auxiliary files.
- Pre-computed model weights or features.
Albumentations provides a mechanism for this using the targets_as_params property.
How it Works:
-
Override
targets_as_params: In your custom transform class, override thetargets_as_paramsproperty. It should return a list of strings. Each string is a key that you expect to find in the input data dictionary passed to theComposepipeline.@property def targets_as_params(self): return ["my_custom_metadata_key", "overlay_image_data"] -
Pass Data to
Compose: When calling the pipeline, include your custom data using the keys defined intargets_as_params.pipeline = A.Compose([ MyCustomTransform(custom_metadata_key="my_custom_metadata_key", ...), ... ]) # Prepare your custom data custom_metadata = {"info": "some_value", "timestamp": 12345} overlay_data = {"image": overlay_img, "mask": overlay_mask} # Pass it during the call result = pipeline( image=main_image, mask=main_mask, my_custom_metadata_key=custom_metadata, # Match the key overlay_image_data=overlay_data # Match the key ) -
Access Data in
get_params_dependent_on_data: The data you passed (associated with the keys listed intargets_as_params) will be available inside thedatadictionary argument of yourget_params_dependent_on_datamethod.def get_params_dependent_on_data(self, params: dict[str, Any], data: dict[str, Any]): # Access standard params like shape height, width = params["shape"] # Access your custom data custom_info = data["my_custom_metadata_key"]["info"] overlay_img = data["overlay_image_data"]["image"] # --- Now use this data to calculate augmentation parameters --- # # Example: Decide blur strength based on custom_info if custom_info == "high_detail": blur_sigma = self.py_random.uniform(0.1, 0.5) else: blur_sigma = self.py_random.uniform(1.0, 2.0) # Example: Process the overlay image (resize, calculate offset, etc.) # (See full OverlayElements example below) processed_overlay_params = self.process_overlay(overlay_img, (height, width)) # Return calculated parameters to be used in apply methods return { "blur_sigma": blur_sigma, "processed_overlay": processed_overlay_params } -
Use Parameters in
apply...Methods: The dictionary returned byget_params_dependent_on_datais passed via**paramsto yourapply,apply_to_mask,apply_to_bboxes, etc., methods.def apply(self, img: np.ndarray, blur_sigma: float, processed_overlay: dict, **params): img = apply_blur(img, blur_sigma) return blend_overlay(img, processed_overlay["image"], processed_overlay["offset"]) def apply_to_mask(self, mask: np.ndarray, processed_overlay: dict, **params): return apply_overlay_mask(mask, processed_overlay["mask"], processed_overlay["mask_id"])
Complete Example: AverageWithExternalImage Transform
The following AverageWithExternalImage transform demonstrates passing an external image via a custom key and averaging it with the main input image.
import albumentations as A
from albumentations.core.transforms_interface import ImageOnlyTransform
import numpy as np
import cv2
from typing import Any, Dict
class AverageWithExternalImage(ImageOnlyTransform):
"""
Averages the input image with an external image passed via `targets_as_params`.
The external image is resized to match the input image dimensions.
"""
def __init__(self, external_image_key: str = "external_image", p: float = 0.5):
"""
Args:
external_image_key (str): The key used to pass the external image data
when calling the pipeline.
p (float): Probability of applying the transform.
"""
super().__init__(p=p)
self.external_image_key = external_image_key
@property
def targets_as_params(self):
"""Specifies that the external image data should be passed as a parameter."""
return [self.external_image_key]
def get_params_dependent_on_data(self, params: dict[str, Any], data: dict[str, Any]):
"""
Retrieves the external image and resizes it to match the target image shape.
"""
if self.external_image_key not in data:
# If the key is missing, return empty params to indicate skipping
return {"resized_external_image": None}
external_image = data[self.external_image_key]
if not isinstance(external_image, np.ndarray):
raise TypeError(f"Expected '{self.external_image_key}' to be a NumPy ndarray, got {type(external_image)}")
target_height, target_width = params["shape"][:2]
# Resize external image to match target image size
resized_external_image = cv2.resize(
external_image, (target_width, target_height), interpolation=cv2.INTER_LINEAR
)
return {"resized_external_image": resized_external_image}
def apply(self, img: np.ndarray, resized_external_image: np.ndarray | None, **params: Any):
"""
Averages the input image with the resized external image.
"""
if resized_external_image is None:
# Skip if external image was not provided or resizing failed
return img
# Ensure both images have the same shape and type for averaging
if img.shape != resized_external_image.shape:
# This might happen if channel numbers differ, handle appropriately
# For simplicity, we'll just return the original image here
# In a real scenario, you might want to convert grayscale to color or vice-versa
print(f"Warning: Shape mismatch between image ({img.shape}) and resized external ({resized_external_image.shape}). Skipping averaging.")
return img
# Perform averaging using float arithmetic for precision, then convert back
avg_image = cv2.addWeighted(img, 0.5, resized_external_image, 0.5, 0.0)
# Ensure the output dtype matches the input dtype
return avg_image.astype(img.dtype)
# --- Example Usage ---
pipeline = A.Compose([
AverageWithExternalImage(external_image_key="reference_photo", p=1.0),
# Other transforms...
])
main_image = cv2.imread("path/to/main_image.jpg")
reference_photo = cv2.imread("path/to/reference_photo.jpg")
augmented_data = pipeline(image=main_image, reference_photo=reference_photo)
averaged_image = augmented_data["image"]
This powerful mechanism allows custom transforms to leverage arbitrary external data or metadata during the augmentation process, enabling highly flexible and complex augmentation scenarios.
Where to Go Next? 🔗
Now that you can create your own custom transforms:
- Integrate Your Transform: Add your custom transform to pipelines within the Basic Usage Guides relevant to your task.
- Explore Base Class APIs: Consult the API Reference for details on
ImageOnlyTransform,DualTransform,Transform3D, and other base classes you might inherit from. - Handle Advanced Scenarios: Learn how custom transforms interact with Additional Targets or how they can be included in Serialization.
- Revisit Core Concepts: Ensure your custom transform correctly handles Targets and fits within the Pipeline structure.
- Contribute (Optional): If your transform is broadly useful, consider contributing it back to the Albumentations library (see the project's contribution guidelines).